Two databases are routinely used to annotate SSU (18S) rRNA eukaryotic metabarcoding data PR2 and Silva. While PR2 is focusing only on eukaryotes (both nuclear and plastid genes), Silva contains both eukaryotes and prokaryotes SSU sequences. Silva tries to be as extensive as possible while PR2 only includes sequences that have been annotated by experts.
Another big difference is that while PR2 uses a fixed number of named taxonomic levels (9 from kingdom to species) and enforces strict taxonomic rules such that a given taxon can only appear in one column, Silva is much more flexible and each sequence can be annotated from 1 to 20 levels which do not have specific names. The advantage of the Silva approach is that taxonomy annotation can be adapted as a function of the groups (e.g. Metazoa groups are often described with many levels such as sub-family, sub-species etc…). However this makes incoherence in taxonomy much harder to track and also makes the analysis of metabarcoding data more complex for example when using packages such as phyloseq that relies on a fixed number of taxonomic levels.
Since version 4.13.0, we provide along with the PR2 annotation the
annotation provided by Silva version 138.1 (field
silva_taxonomy which will look typically as string
separated by “;”. For example:
- “Eukaryota;SAR;Rhizaria;Cercozoa;Thecofilosea;Ebriacea;Ebria;uncultured eukaryote”
- “Eukaryota;Amorphea;Obazoa;Opisthokonta;Holozoa;Choanozoa;Metazoa;Animalia;BCP clade;Bilateria;Deuterostomia;Chordata;Tunicata;Ascidiacea;Enterogona;Ciona savignyi”
In this small tutorial we offer some hints on how to compare the annotations between the two databases.
# Loading the necessary packages
library("ggplot2")
library("dplyr")
library("tidyr")
library("DT")
library("forcats")
library("stringr")
library("treemapify")
library(patchwork)
library("pr2database")
packageVersion("pr2database")
#> [1] '5.1.0'
# Read the PR2 database
pr2 <- pr2_database() %>%
# Only keep 18S (do not consider plastids)
filter(gene == "18S_rRNA")Structure of Silva annotations
# Split the field silva_taxonomy into individual columns
taxo_ranks_pr2 = c("domain", "supergroup", "division", "subdivision", "class", "order",
"family", "genus", "species")
taxo_ranks_silva = str_c("rank_silva_", 1:20)
pr2_silva <- pr2 %>%
select(pr2_accession, domain:species, silva_taxonomy) %>%
# Remove sequences for which we have no taxonomy from Silva
filter(!is.na(silva_taxonomy)) %>%
# Count the number of Silva taxonomy levels for each sequence
mutate(silva_levels = str_count(silva_taxonomy, ";") + 1) %>%
# Split Silva taxonomy into 20 individuals columns
separate(col = silva_taxonomy, into = taxo_ranks_silva,
sep = ";", remove = FALSE)
Total number of PR2 sequences with Silva annotation : 173003
In contrast to PR2, Silva does not have a fixed number of taxonomy levels. The maximum number of levels is 20. We compute the number of number of levels for each sequences and for each level the number of distinct names in that column
# Plot number of levels for each sequence
g1 <- ggplot(pr2_silva) +
geom_bar(aes(x = silva_levels, fill = as.factor(silva_levels))) +
labs(title = "Silva annotations uses a variable number of levels",
y = "Number of sequences",
x = "Number of annotations levels in Silva") +
theme_classic() +
scale_fill_viridis_d() +
guides(fill=FALSE)
# Compute and plot number of different names for each level of silva (column)
pr2_silva_taxa <- pr2_silva %>%
select(rank_silva_1:rank_silva_20) %>%
summarise_all(~ n_distinct(.)) %>%
pivot_longer(cols = contains("silva"),
names_to = "level",
values_to = "n_taxa_names") %>%
mutate(level = as.integer(str_replace(level, "rank_silva_", "")))
g2 <- ggplot(pr2_silva_taxa) +
geom_col(aes(x = level, y = n_taxa_names, fill = level)) +
labs(title = "Number of different taxonomic names for each annotation level",
y = "Number of different of taxa",
x = "Annotation level (or column) in Silva") +
theme_classic() +
scale_fill_viridis_c() +
guides(fill=FALSE)
g1 / g2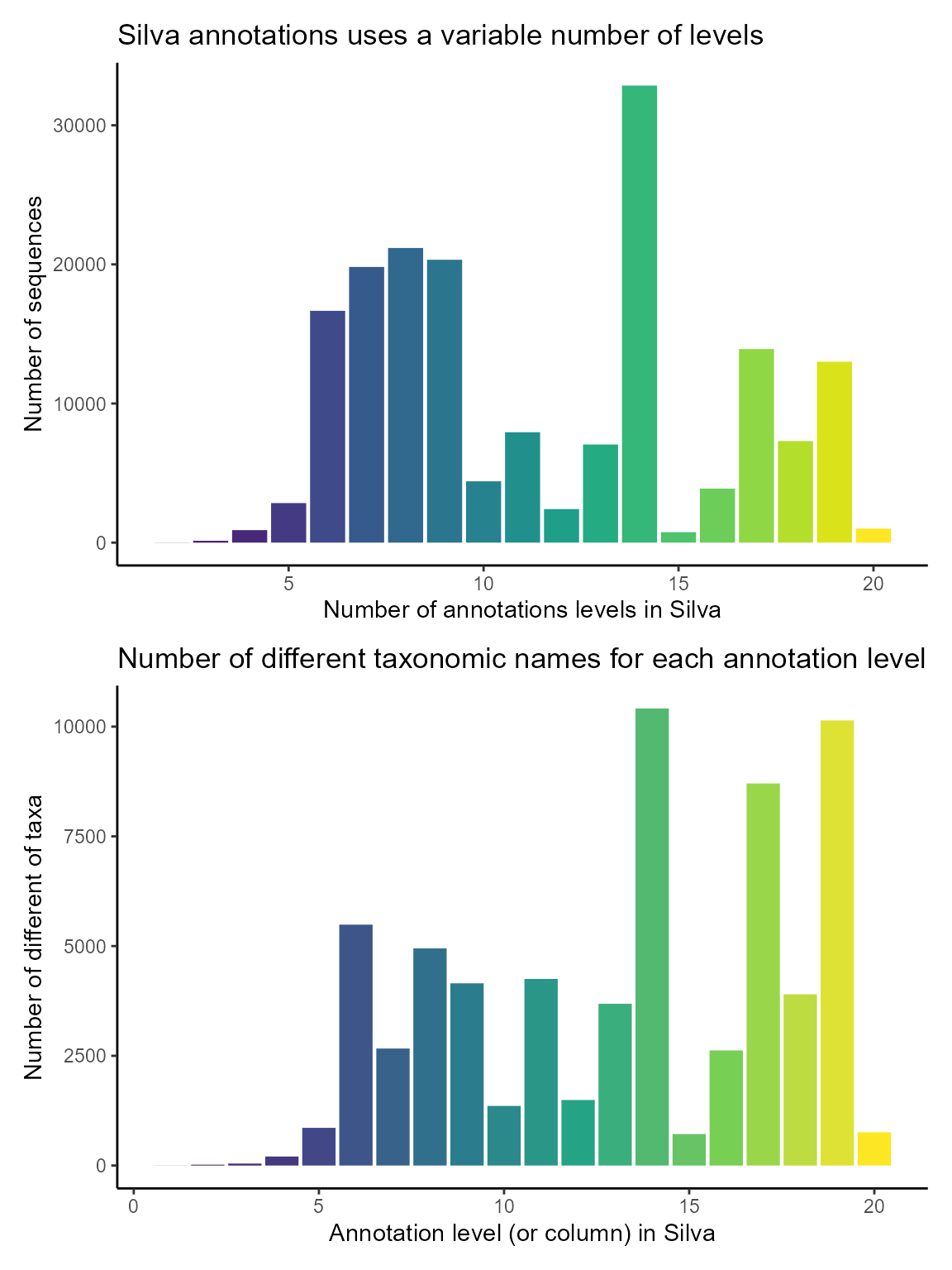
Comparison of taxonomic composition between PR2 and Silva
# Define a function for treemaps
pr2_treemap <- function(pr2, level1, level2) {
# Group
pr2_class <- pr2 %>%
count({{level1}},{{level2}}) %>%
filter(!is.na({{level2}})) %>%
ungroup()
# Do a treemap
ggplot(pr2_class, aes(area = n, fill = {{level2}},
subgroup = {{level1}}, label = {{level2}})) +
treemapify::geom_treemap()
ggplot(pr2_class, aes(area = n, fill= {{level1}},
subgroup = {{level1}}, label = {{level2}})) +
treemapify::geom_treemap() +
treemapify::geom_treemap_text(colour = "white",
place = "centre", grow = TRUE) +
treemapify::geom_treemap_subgroup_border() +
treemapify::geom_treemap_subgroup_text(place = "centre", grow = T,
alpha = 0.5, colour = "black",
min.size = 0) +
theme_bw() +
scale_color_brewer() +
guides(fill = FALSE)
}PR2 supergroup level
Silva level 3 corresponds roughly to PR2 level 2 (supergroup). But this is not true for all taxa.
g1 <- pr2_treemap(pr2_silva, supergroup, division) +
labs(title = "PR2 - Supergroup")
g2 <- pr2_treemap(pr2_silva, rank_silva_3, rank_silva_4) +
labs(title = "Silva - level 3")
g1+ g2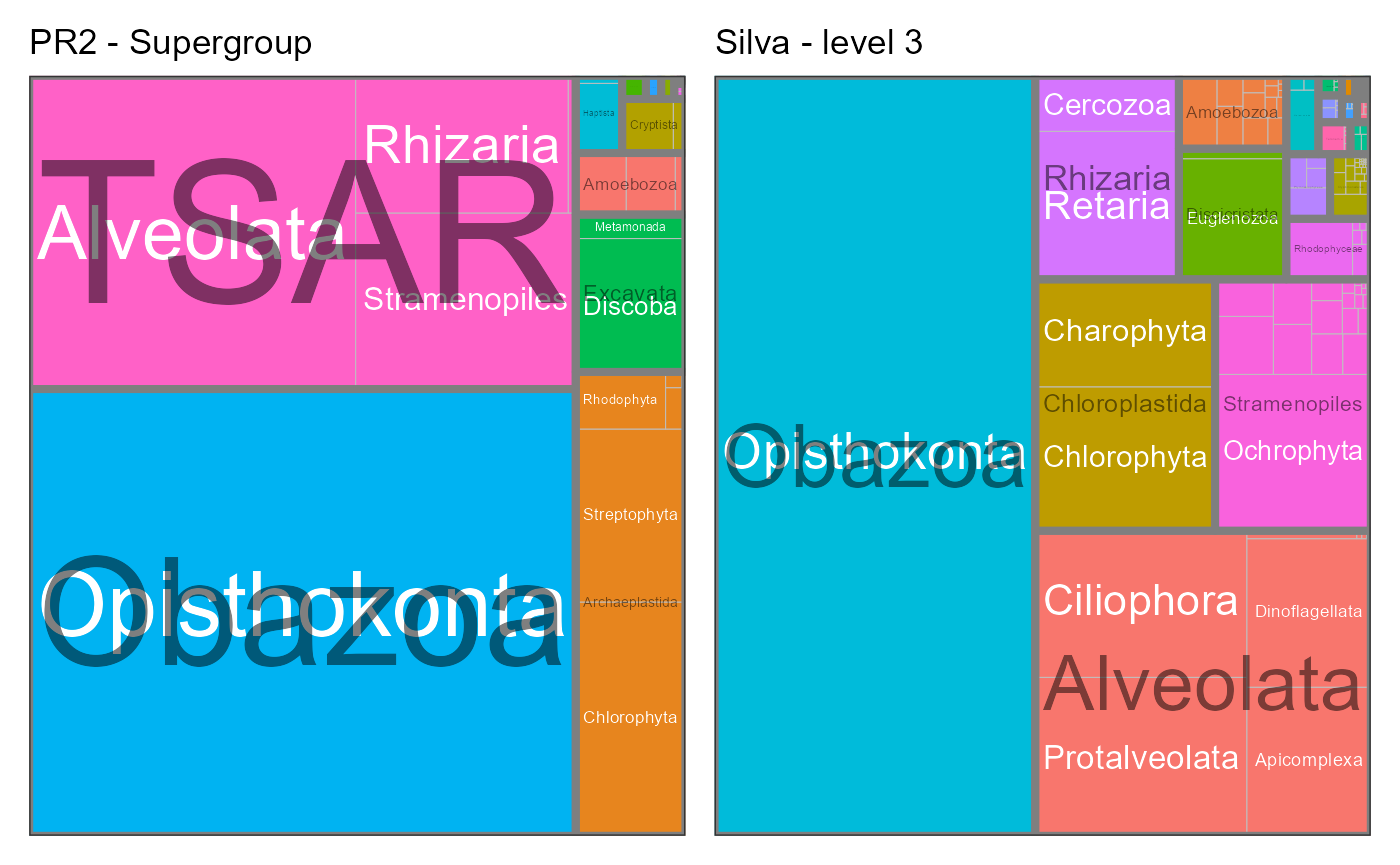
Some issues with Silva annotations
Many annotations contain “uncultured”
There is no really coherent use of “uncultured” in Silva annotations as “uncultured” can appear at one level and then the next level down correspond to a described taxon.
table <- pr2_silva %>%
filter(str_detect(silva_taxonomy, "uncultured")) Number of sequences annotated by Silva as “uncultured”:
rnrow(table)`
We will be taking one example from the Spirotrichea (Ciliophora).
table <- table %>%
filter(class == "Spirotrichea") %>%
select(pr2_accession, order:species, rank_silva_7:rank_silva_9 ) %>%
arrange(order, family, genus, species)
DT::datatable(table,
caption="Spirotrichea (division= Ciliophora) taxa annotated as uncultured in Silva. There is no real coherence in the number of ranks provided by Silva.",
rownames = FALSE)Species for which higher ranks are not annotated
As an example, we extracted Alveolata sequences that have from 1 to 4 levels of annotations according to Silva. As you can see, some are only annotated with the supergroup and the species but all other ranks are missing while in PR2 all levels are clearly annotated.
table <- pr2_silva %>%
filter(silva_levels <=4) %>%
filter(!str_detect(silva_taxonomy, "uncultured"))%>%
filter(division == "Alveolata") %>%
select(pr2_accession, rank_silva_2:rank_silva_4, class:species)
DT::datatable(table,
caption="Alveolata sequences with only 4 levels annotated in Silva",
rownames = FALSE)