library("ggplot2")
library("dplyr")
library("knitr")
library("forcats")
library("stringr")
library("rworldmap")
library("treemapify")
library("pr2database")
packageVersion("pr2database")
#> [1] '5.1.0'
pr2 <- pr2_database()
pr2_photo <- pr2 %>%
filter((division %in% c("Chlorophyta", "Dinophyta",
"Cryptophyta",
"Haptophyta", "Ochrophyta")) &
!(class %in% c("Syndiniales", "Sarcomonadea")))
pr2_ref <- pr2 %>% filter(!is.na(reference_sequence))PR2 fields
colnames(pr2)
#> [1] "pr2_main_id" "pr2_accession"
#> [3] "domain" "supergroup"
#> [5] "division" "subdivision"
#> [7] "class" "order"
#> [9] "family" "genus"
#> [11] "species" "genbank_accession"
#> [13] "start" "end"
#> [15] "label" "gene"
#> [17] "organelle" "taxo_id"
#> [19] "species_version_5.0" "reference_sequence"
#> [21] "remark" "mixoplankton"
#> [23] "HAB_species" "ecological_function"
#> [25] "worms_id" "worms_marine"
#> [27] "worms_brackish" "worms_freshwater"
#> [29] "worms_terrestrial" "gbif_id"
#> [31] "sequence" "sequence_length"
#> [33] "ambiguities" "sequence_hash"
#> [35] "gb_date" "gb_division"
#> [37] "gb_definition" "gb_organism"
#> [39] "gb_organelle" "gb_taxonomy"
#> [41] "gb_strain" "gb_culture_collection"
#> [43] "gb_clone" "gb_isolate"
#> [45] "gb_isolation_source" "gb_specimen_voucher"
#> [47] "gb_host" "gb_collection_date"
#> [49] "gb_environmental_sample" "gb_country"
#> [51] "gb_lat_lon" "gb_collected_by"
#> [53] "gb_note" "gb_sequence"
#> [55] "gb_publication" "gb_authors"
#> [57] "gb_journal" "eukref_name"
#> [59] "eukref_source" "eukref_env_material"
#> [61] "eukref_env_biome" "eukref_biotic_relationship"
#> [63] "eukref_specific_host" "eukref_geo_loc_name"
#> [65] "eukref_notes" "pr2_sample_type"
#> [67] "pr2_sample_method" "pr2_latitude"
#> [69] "pr2_longitude" "pr2_depth"
#> [71] "pr2_ocean" "pr2_sea"
#> [73] "pr2_sea_lat" "pr2_sea_lon"
#> [75] "pr2_country" "pr2_location"
#> [77] "pr2_location_geoname" "pr2_location_geotype"
#> [79] "pr2_location_lat" "pr2_location_lon"
#> [81] "pr2_sequence_origin" "pr2_size_fraction"
#> [83] "metadata_remark" "pr2_continent"
#> [85] "pr2_country_geocode" "pr2_country_lat"
#> [87] "pr2_country_lon" "eukribo_UniEuk_taxonomy_string"
#> [89] "eukribo_V4" "eukribo_V9"
#> [91] "silva_taxonomy" "organelle_code"
#> [93] "species_rod" "infraspecific_name"
#> [95] "isolate" "assembly_level"
#> [97] "assembly_type" "gc_percent"
#> [99] "pubmed_id" "species_url"
#> [101] "accession_genbank_link"Basic statistics
All taxa
Total number of PR2 sequences : 240200
pr2_taxa <- pr2 %>% select(domain:genus, species) %>%
summarise_all(funs(n_distinct(.)))
knitr::kable(pr2_taxa, caption="Number of taxa - all sequences")| domain | supergroup | division | subdivision | class | order | family | genus | species |
|---|---|---|---|---|---|---|---|---|
| 8 | 39 | 105 | 140 | 449 | 1136 | 2423 | 26266 | 54123 |
Photosynthetic protists
Number of photosynthetic protist sequences : 15412
pr2_taxa <- pr2_photo %>% select(domain:genus, species) %>%
summarise_all(funs(n_distinct(.)))
knitr::kable(pr2_taxa, caption="Number of taxa - photosynthetic protist sequences")| domain | supergroup | division | subdivision | class | order | family | genus | species |
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 3 | 3 | 24 | 62 | 94 | 516 | 1822 |
Reference sequences
Reference sequences are a subset of PR2 representative of taxonomic groups.
Number of reference sequences : 240200
pr2_taxa <- pr2_ref %>% select(domain:genus, species) %>%
summarise_all(funs(n_distinct(.)))
knitr::kable(pr2_taxa, caption="Number of taxa - Reference sequences")| domain | supergroup | division | subdivision | class | order | family | genus | species |
|---|---|---|---|---|---|---|---|---|
| 8 | 39 | 105 | 140 | 449 | 1136 | 2423 | 26266 | 54123 |
Sequence length
ggplot(pr2) + geom_histogram(aes(sequence_length), binwidth = 100, fill="blue") +
xlim(0,3000) + xlab("PR2 sequence length") +
ylab("Number of sequences") +
ggtitle("All sequences")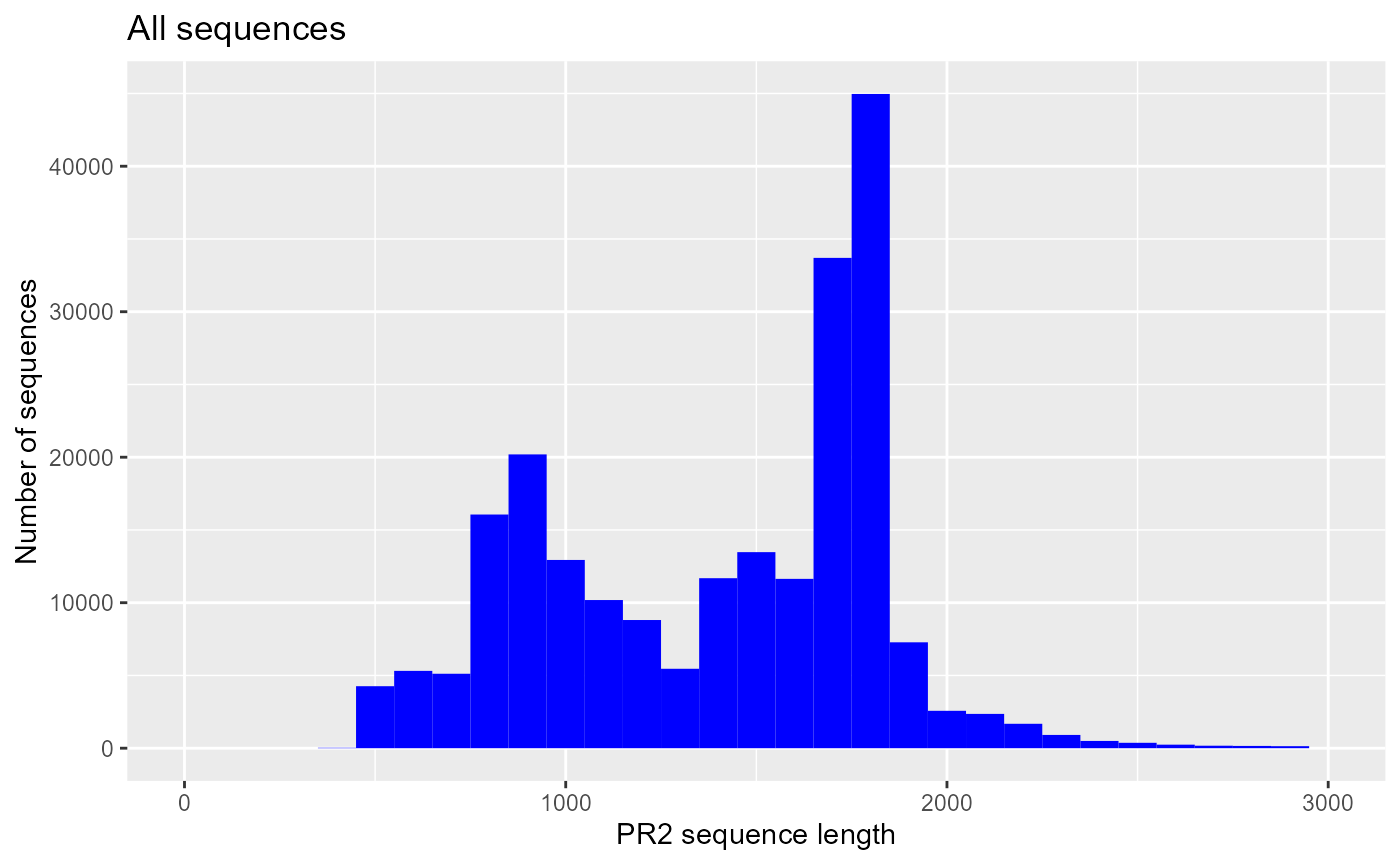
ggplot(pr2_ref) + geom_histogram(aes(sequence_length), binwidth = 100, fill="blue") +
xlim(0,3000) +
xlab("PR2 sequence length") +
ylab("Number of sequences") +
ggtitle("Reference sequences")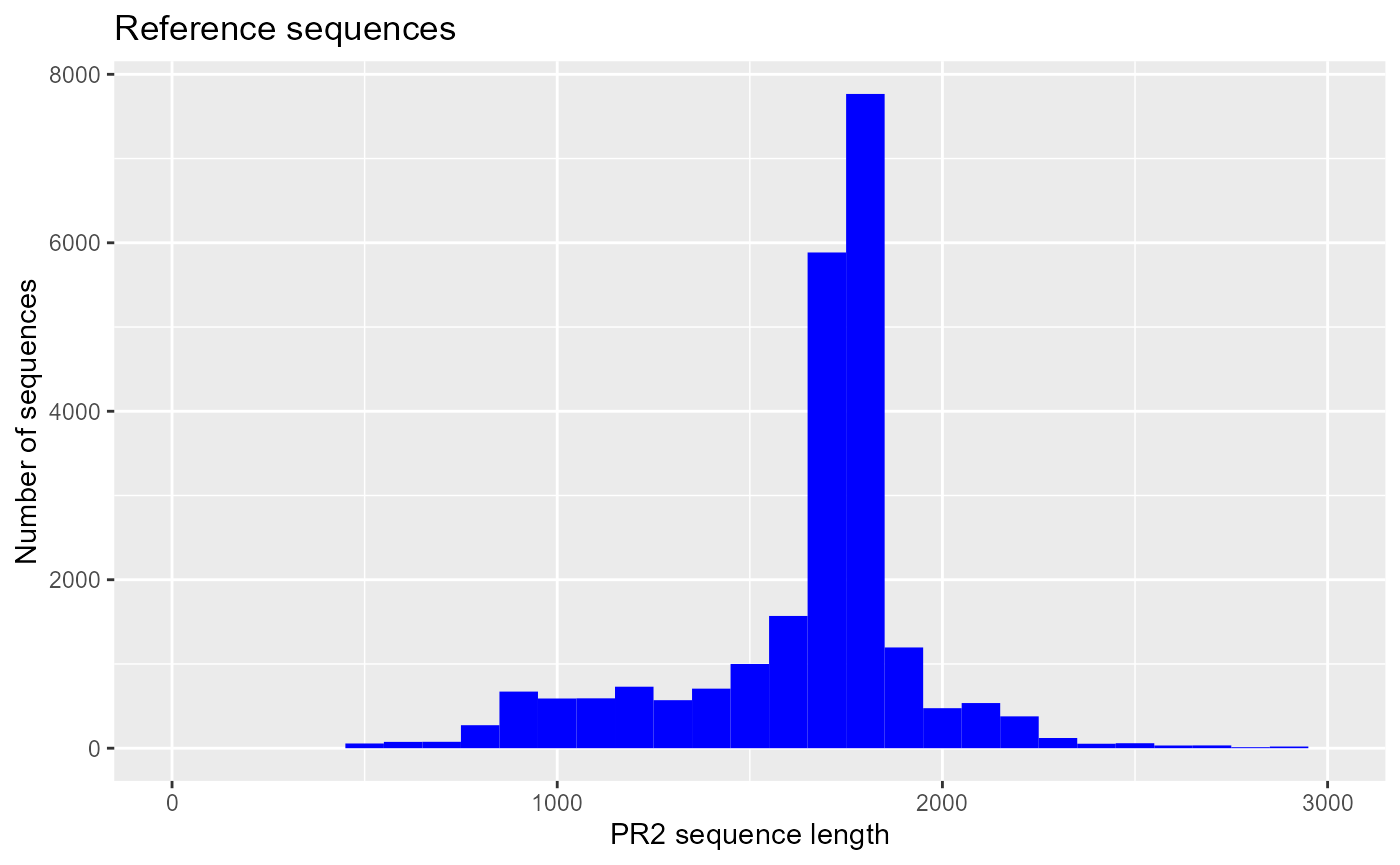
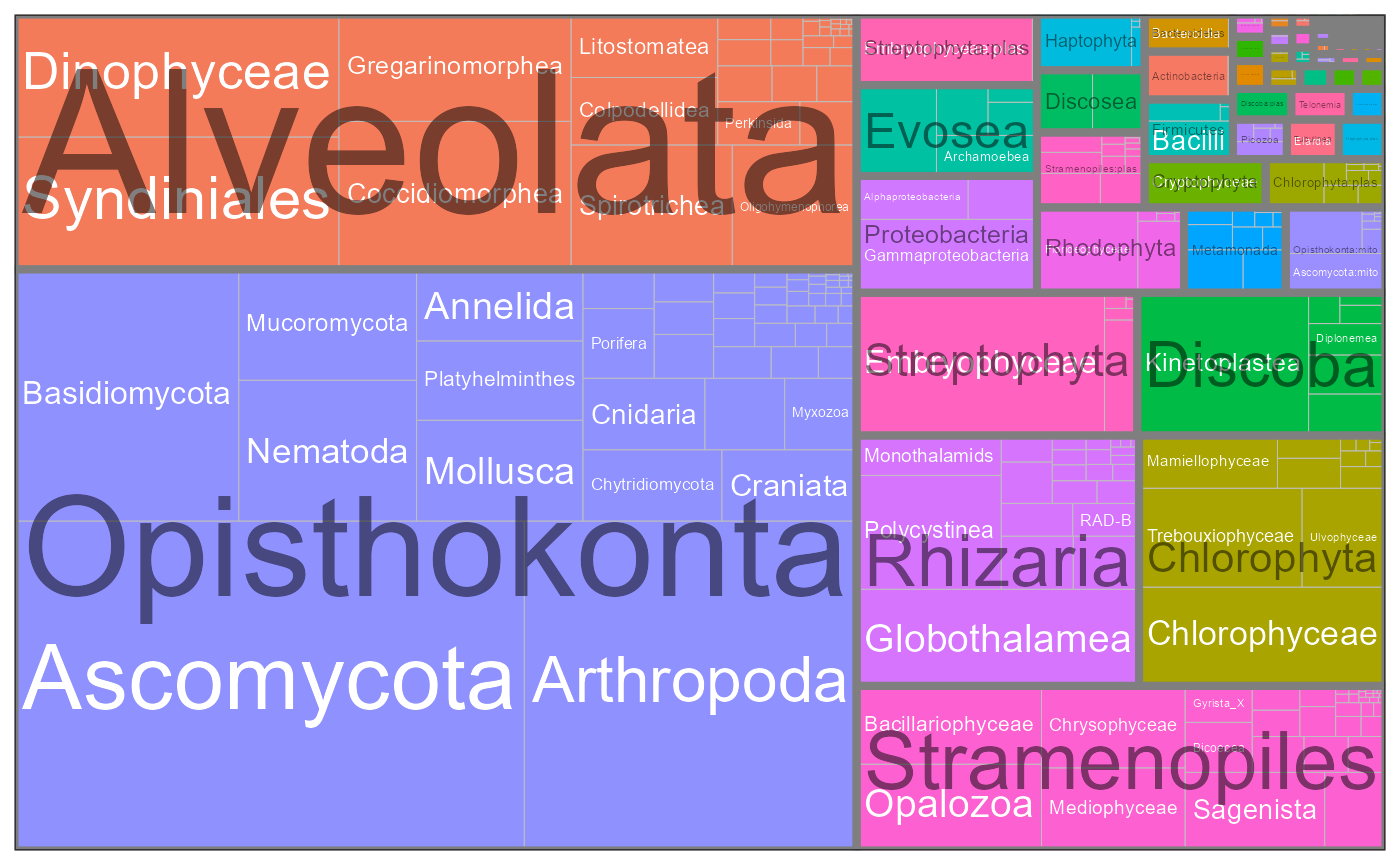
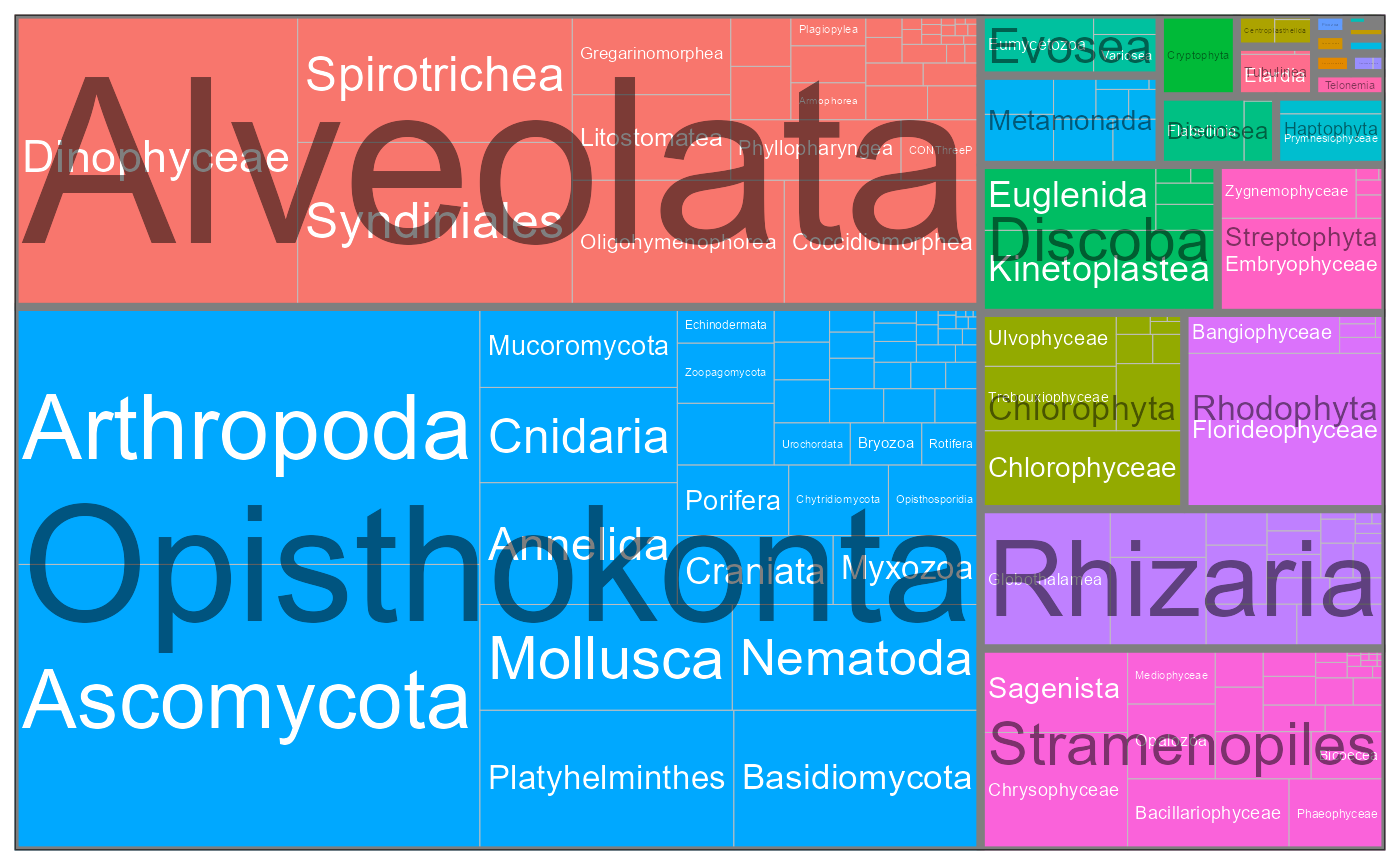
Taxonomic composition
pr2_treemap <- function(pr2, level1, level2) {
# Group
pr2_class <- pr2 %>%
count({{level1}},{{level2}}) %>%
filter(!is.na(division)) %>%
ungroup()
# Do a treemap
ggplot(pr2_class, aes(area = n,
fill = {{level2}},
subgroup = {{level1}},
label = {{level2}})) +
treemapify::geom_treemap()
ggplot(pr2_class, aes(area = n,
fill= {{level1}},
subgroup = {{level1}},
label = {{level2}})) +
treemapify::geom_treemap() +
treemapify::geom_treemap_text(colour = "white", place = "centre", grow = TRUE) +
treemapify::geom_treemap_subgroup_border() +
treemapify::geom_treemap_subgroup_text(place = "centre", grow = T,
alpha = 0.5, colour = "black",
min.size = 0) +
theme_bw() +
scale_color_brewer() +
guides(fill = FALSE)
}Genera most represented
All taxa
pr2_genus <- pr2 %>% group_by(class, genus) %>%
count() %>%
ungroup() %>%
top_n(30)
ggplot(pr2_genus) +
geom_col(aes(x=forcats::fct_reorder(stringr::str_c(class,"-",genus), n), y=n)) +
coord_flip() +
ggtitle("Most represented genera - all") +
xlab("Genera") + ylab("Number of sequences")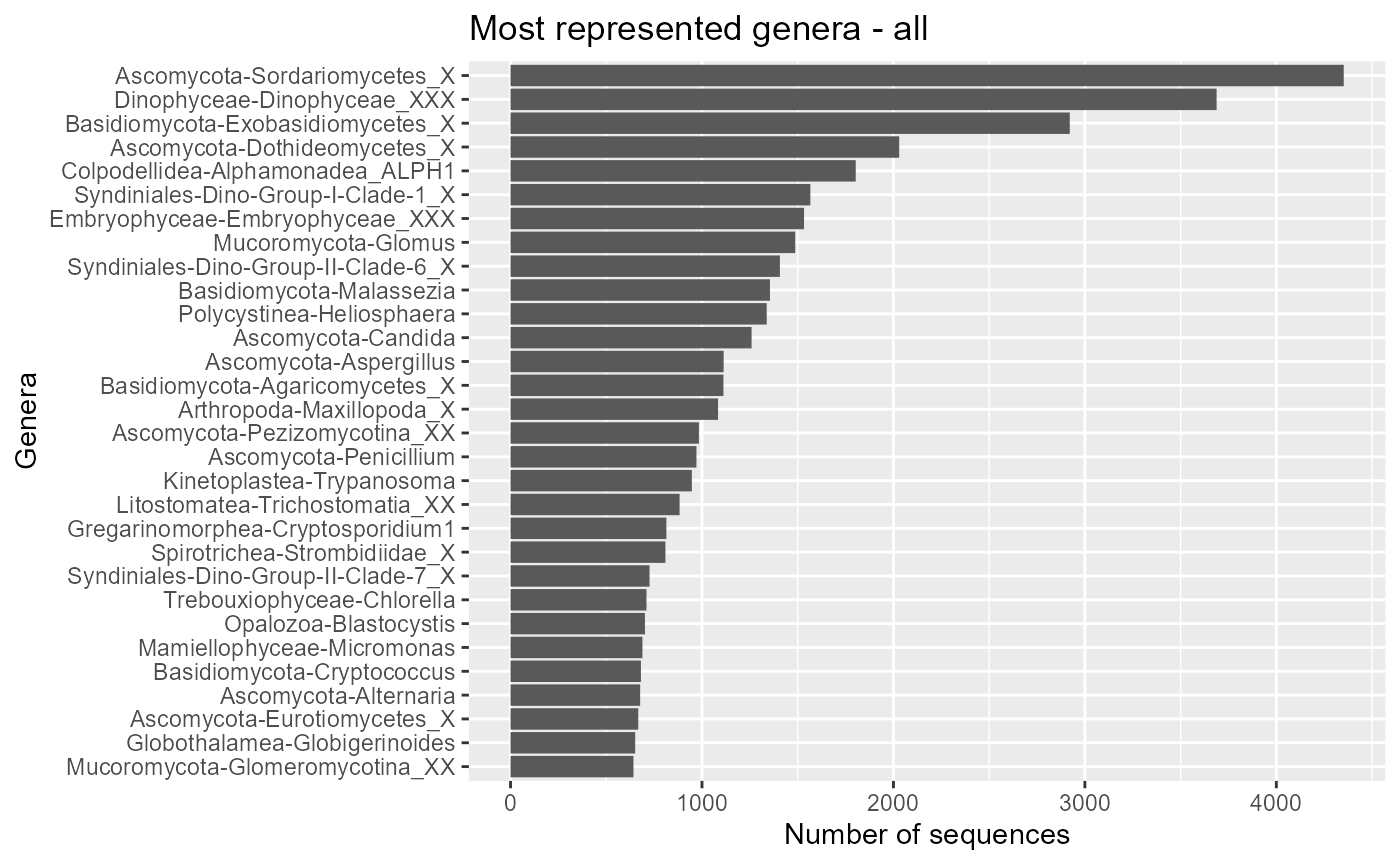
Reference sequences
pr2_genus <- pr2_ref %>%
group_by(class, genus) %>%
count() %>% ungroup() %>%
top_n(30)
ggplot(pr2_genus) +
geom_col(aes(x=forcats::fct_reorder(stringr::str_c(class,"-",genus), n), y=n)) +
coord_flip() +
ggtitle("Reference sequences") +
xlab("Genera") +
ylab("Number of sequences")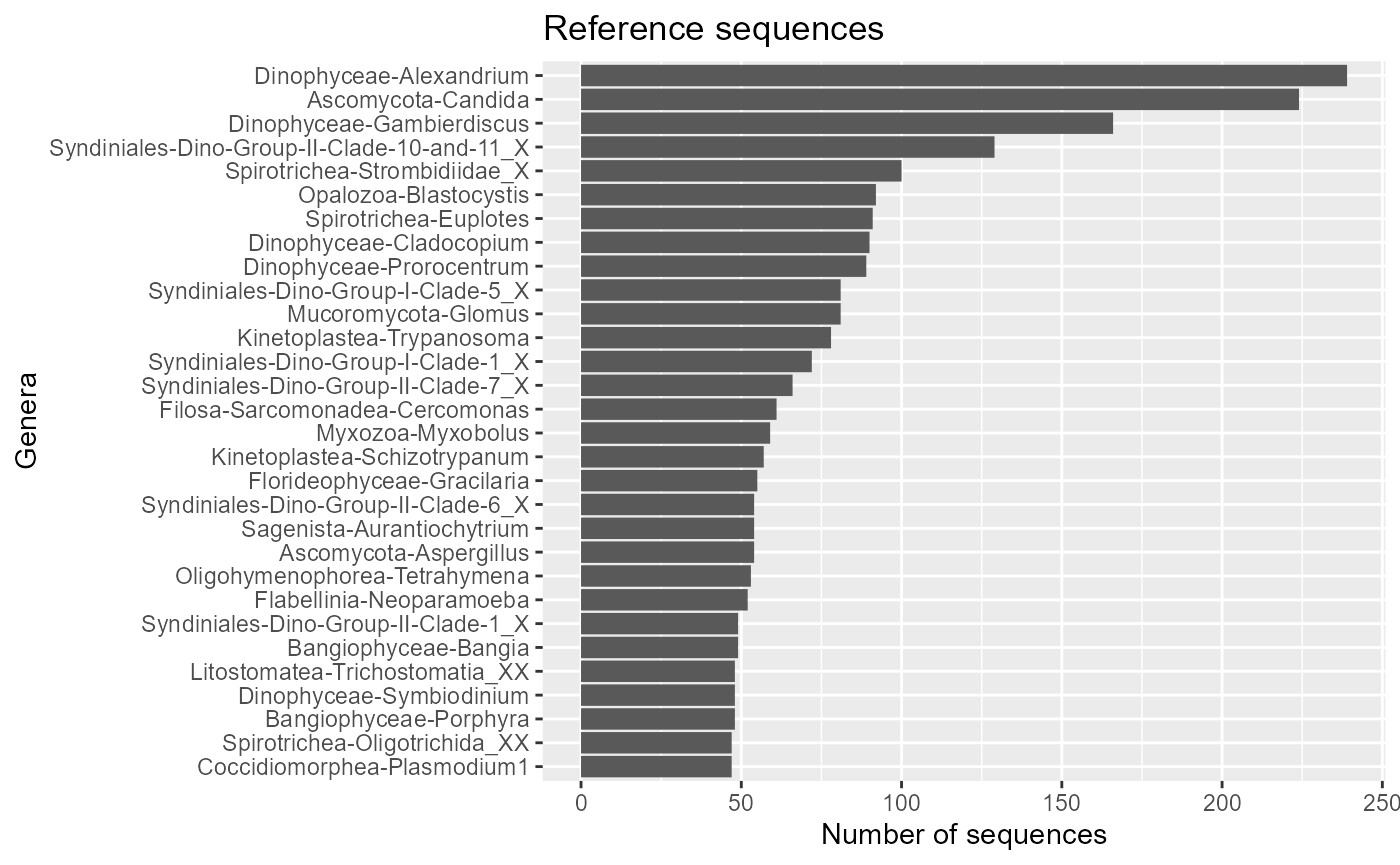
Only photosynthetic protists
pr2_genus <- pr2_photo %>%
group_by(class, genus) %>%
count() %>%
ungroup() %>%
top_n(30)
ggplot(pr2_genus) +
geom_col(aes(x=forcats::fct_reorder(stringr::str_c(class,"-",genus), n), y=n)) +
coord_flip() +
ggtitle("Most represented genera - only photosynthetic protists") +
xlab("Genera") + ylab("Number of sequences")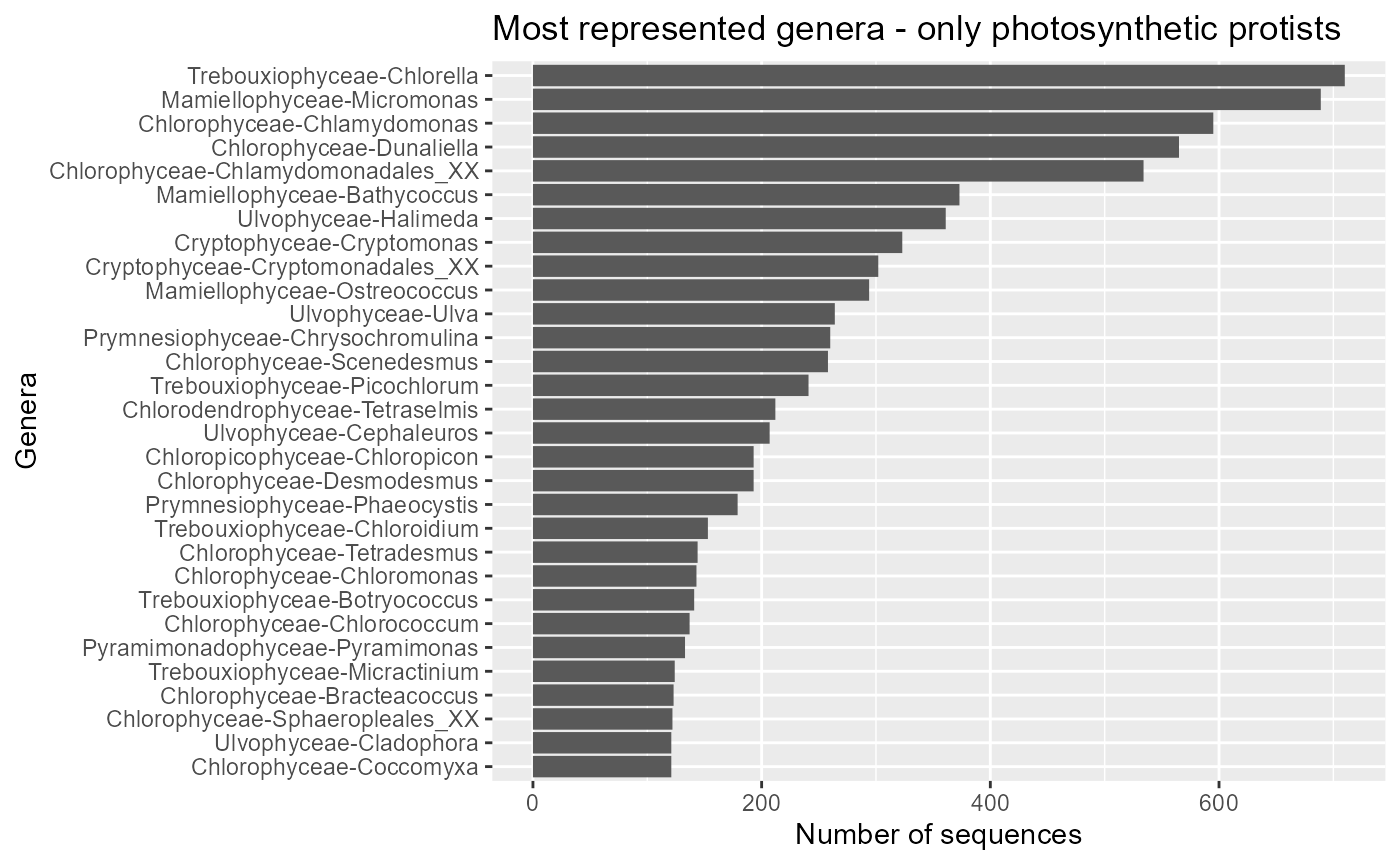
World sequence distribution
map_get_world <- function(resolution="coarse"){
# Change to "coarse" for global maps / "low" for regional maps
worldMap <- rworldmap::getMap(resolution = resolution)
world.points <- fortify(worldMap)
world.points$region <- world.points$id
world.df <- world.points[,c("long","lat","group", "region")]
}
map_world <- function(color_continents = "grey80",
color_borders = "white",
resolution = "coarse") {
# Background map using the maps package
# world.df <- map_data("world")
world.df <- map_get_world(resolution)
map <- ggplot() +
geom_polygon(data = world.df,
aes(x=long, y = lat, group = group),
fill=color_continents,
color=color_borders) +
# scale_fill_manual(values= color_continents , guide = FALSE) +
scale_x_continuous(breaks = (-4:4) * 45) +
scale_y_continuous(breaks = (-2:2) * 30) +
xlab("Longitude") + ylab("Latitude") +
coord_fixed(1.3) +
theme_bw()
# species_map <- species_map + coord_map () # Mercator projection
# species_map <- species_map + coord_map("gilbert") # Nice for the poles
return(map)
}All taxa
map_world() + geom_point(data=pr2, aes(x=pr2_longitude, y=pr2_latitude),
fill="blue", size=2, shape=21) +
ggtitle("PR2 - all sequences") 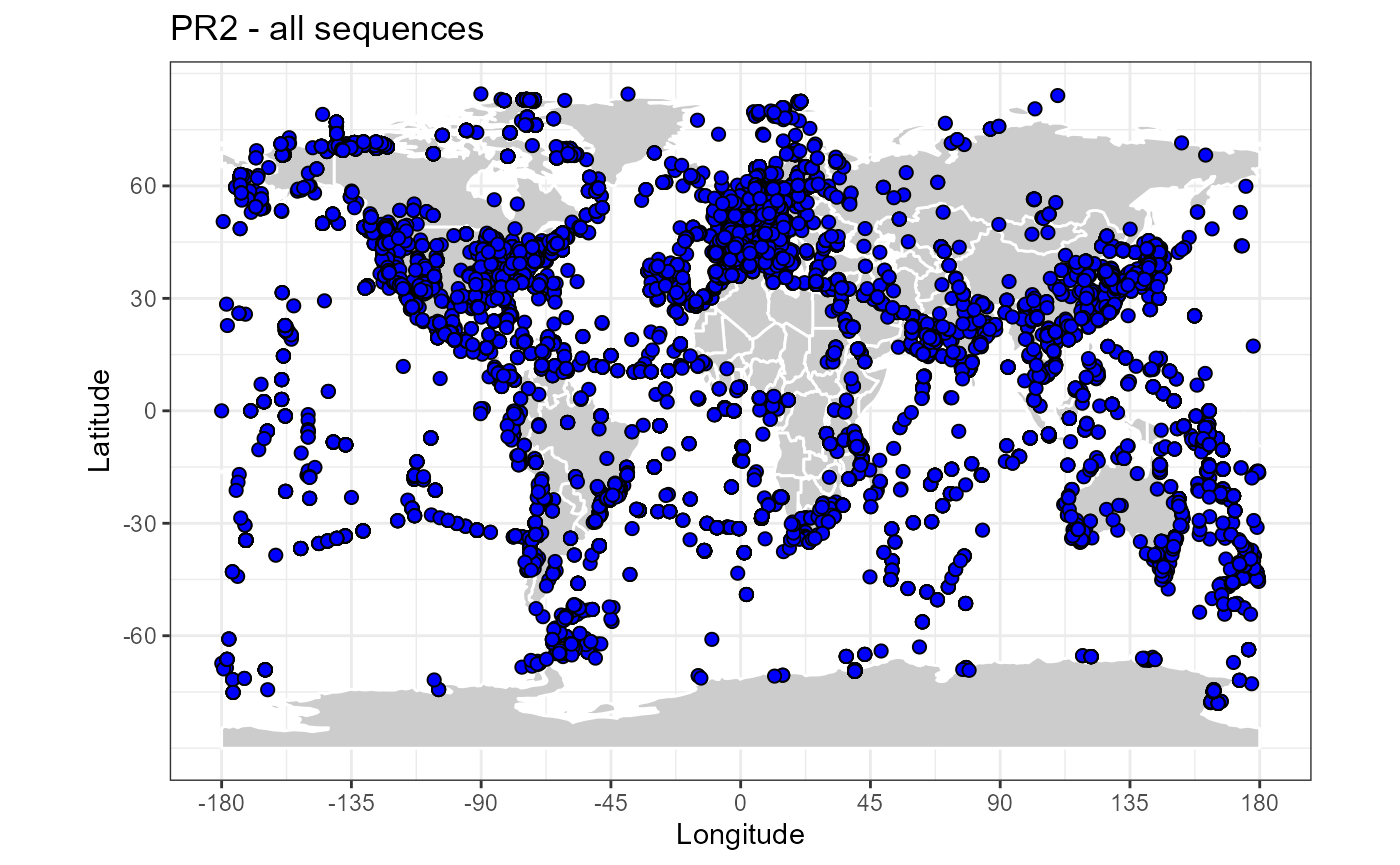
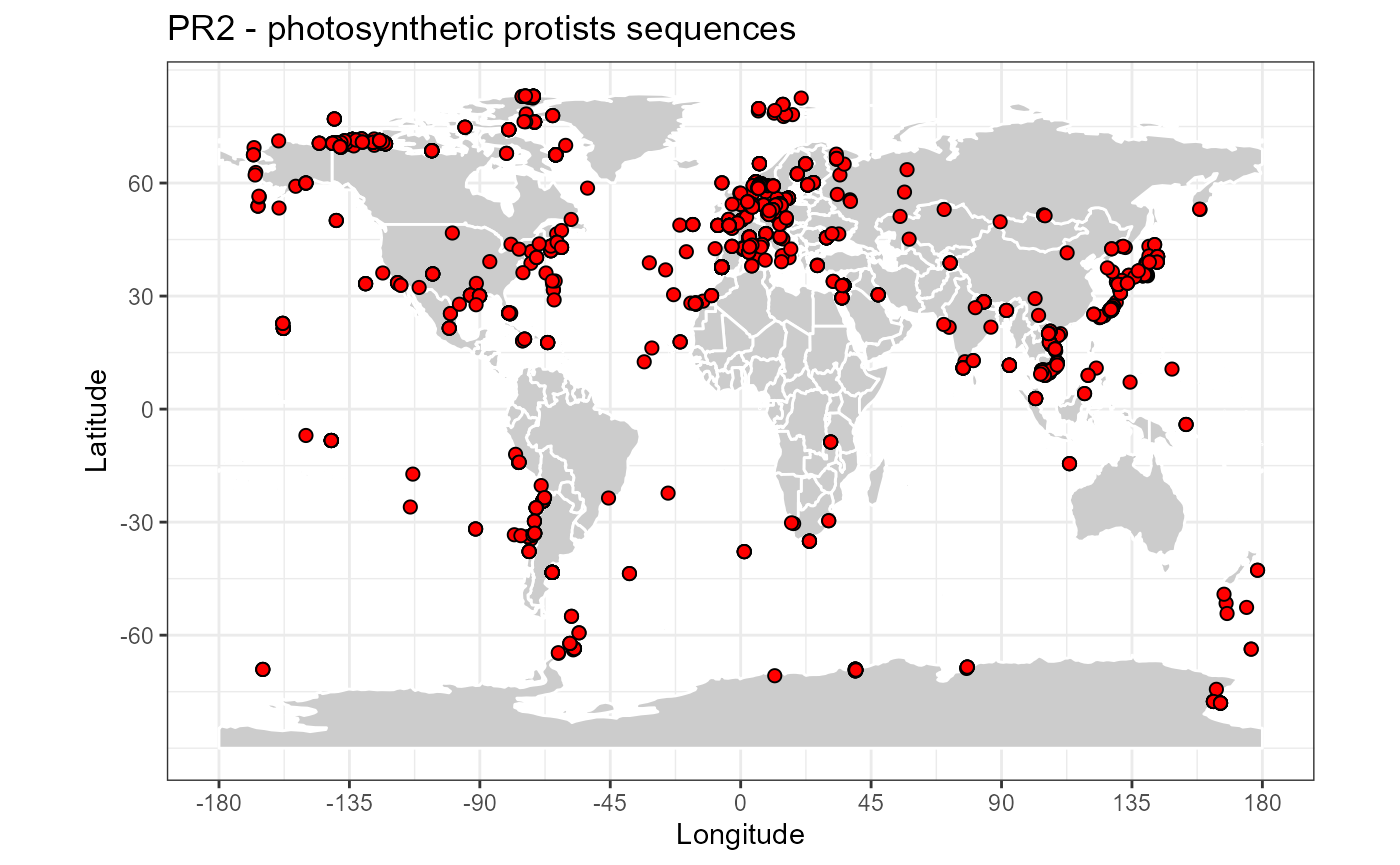
Photosynthetic protists
map_world() + geom_point(data=pr2_photo, aes(x=pr2_longitude, y=pr2_latitude),
fill="red", size=2, shape=21) +
ggtitle("PR2 - photosynthetic protists sequences")